“Our group is eclectic,” said Borchers. “It’s a bunch of people with
computational backgrounds in different bioscience fields. We spend less
time processing data and more time developing the approaches and methods
to address the questions that need answering for projects that are
computational in nature.”
During his internship, Borchers developed a computational method to
estimate the size of human centromeres, which he brought to the T2T
project.
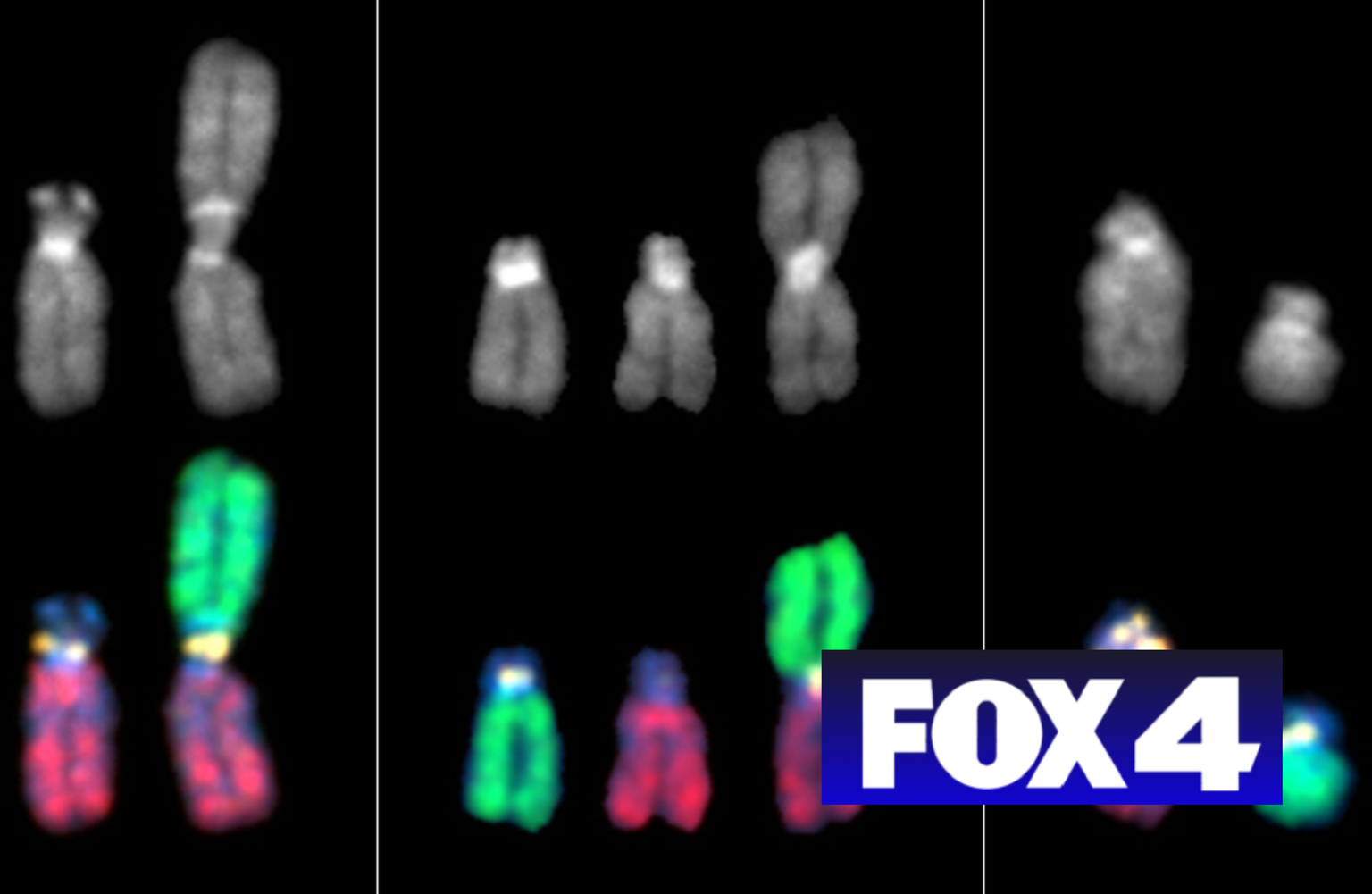
“We compared the newly-assembled centromeric sequences from the X
chromosome of the CHM13 cell line with those from the 1000 Genomes
Project, which had samples from different individuals from all over the
world, to see how CHM13 compared to a more diverse representation of
centromere sequences,” said Borchers.
Consistent with previous research, they found substantial size
variation in centromeres between individuals with different geographic
ancestry. The researchers published their work in a third Science paper,
led by Nick Altemose, PhD, and Miga, reporting the characterization of
the genetic and epigenetic landscape of human centromeres.
Productive during the pandemic
Initial efforts from the T2T consortium resulted in the complete
assembly of chromosomes X and 8, coauthored by Stowers researchers and
published in 2020 and 2021 in Nature. It has since grown into a team of about 100 talented individuals, mostly computational biologists.
“In twenty years from now, when we look back on this human genome
project 2.0, it will become clear that it really happened during the
pandemic,” reflected Gerton.
Gerton, Potapova, and Gomes de Lima all gave credit to Miga and Phillippy, the two co-leaders of the T2T consortium.
“They established some ground rules early on about how they wanted
the consortium to work. It’s a public good we are generating, and they
really want people to be collaborative and work together,” said Gerton.