Research
Research
Codon Optimality in Vertebrates
We seek to understand what dictates the stability and the level of translation of messenger RNA (mRNA) in vertebrates. In particular, we are decoding the regulatory information encrypted across the ENTIRE mRNA. And while we are interested in the canonical regulatory mechanism by which proteins (e.g. RNA binding proteins) or microRNAs recognize regulatory elements in the 3’UTRs, we are also very interested in the regulatory information encrypted in the coding region, and more precisely, in the genetic code. We have demonstrated that mRNA translation strongly affects mRNA stability in a codon-dependent manner. In sum, we seek to understand how mRNA stability and translation is regulated during embryogenesis and how that may affect human diseases.
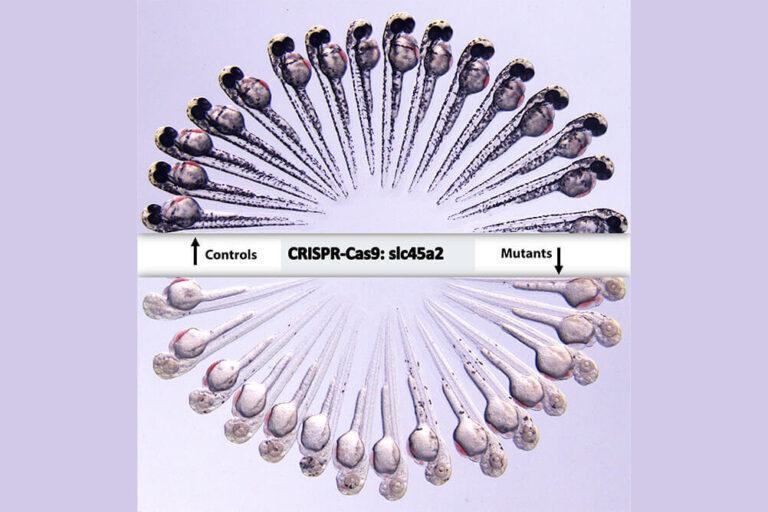
CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos
We demonstrated that CRISPR-RfxCas13d is an effective and precise system to deplete specific mRNA transcripts in zebrafish embryos. Both zygotically-expressed and maternally-provided transcripts can be efficiently targeted in zebrafish embryos, resulting in a 75% average decrease in transcript levels and generate developmental phenotypes. Moreover, CRISPR-RfxCas13d can be used in medaka, killifish and mouse embryos.
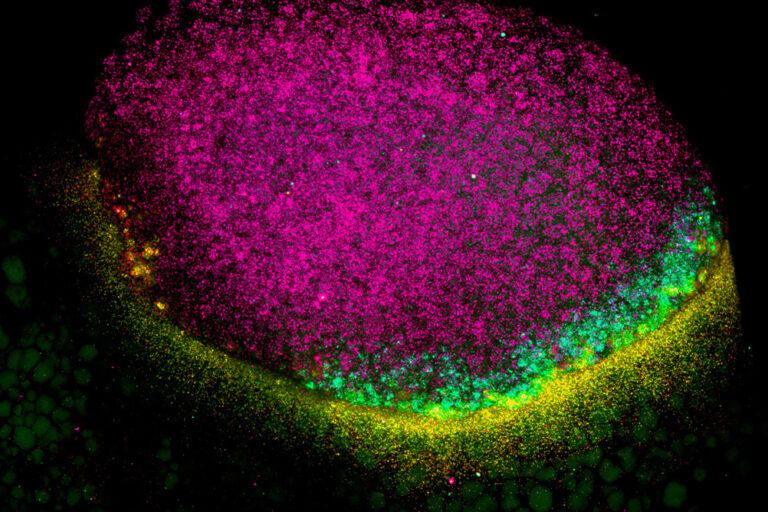
Downstream ORF
The assumption that mRNAs in higher organisms translate a single ORF has undergone a dramatic revision in recent years. Thousands of small translated ORFs have been identified within previously assigned UTRs and long non-coding RNAs. Translation of small ORFs in the 5’UTR, known as upstream-ORFs, has been widely studied and generally tend to attenuate translation of the associated main ORF. We have discovered translated small ORFs in the 3’UTR, which we refer to as downstream open reading frames (dORFs). Unexpectedly, we found that these dORFs strongly enhance translation of the main coding ORF. We are interesting in dissect the dORF molecular mechanism and biological role in development and relation with human diseases.
Codon Optimality
CRISPR-Cas13d
dORF
The Bazzini Lab uncovers fundamental rules for how dengue virus infects its mosquito and human hosts, providing hope for identifying therapeutic approaches.
Horacio Pallares, a postdoc in the Bazzini Lab, discusses his zika virus research.
Bazzini Lab Predoctoral Researcher Luciana Castellano explains how understand the genetic code.