Research
Our Research
How can we navigate vast sequence spaces to decode genetic behavior?
Research areas: Genetics and Genomics, Molecular and Cell Biology, Systems Biology
Research in the Garruss Lab
Our research explores the integration of massively multiplexed experimentation and machine learning to understand fundamental properties of gene expression and behavior. The transcription and translation of genetic information play critical roles in all biological systems yet grand mysteries remain about how the primary sequence of DNA, RNA, and proteins affect genetic outcomes. We develop innovative approaches to decode sequence-function relationships to improve our understanding of core genetic systems and to unlock applications in healthcare and environmental sustainability.
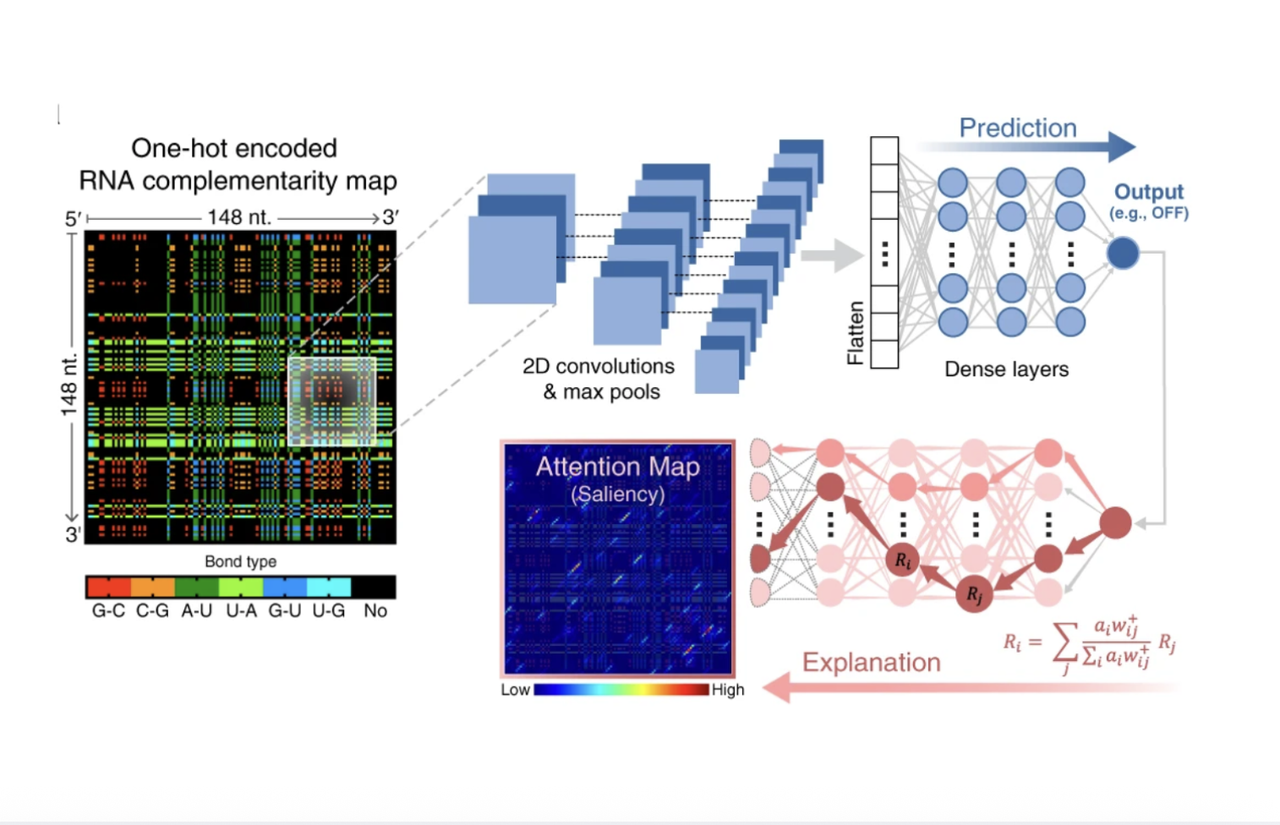
Our prior work established programmable translation control based on RNA toehold switches that conditionally block translation of a gene transcript, only allowing protein expression if a separate trigger RNA is detected. Toehold switches allow for the live, intracellular biosensing of endogenous RNA levels as well as rapid detection of pathogens/infections. We generated and experimentally tested ~100,000 toehold-trigger pairs to discover the RNA sequence- and structure-based design rules behind the most effective switches. We used convolutional neural networks trained on features of RNA secondary structure to visualize and understand success and failure modes.
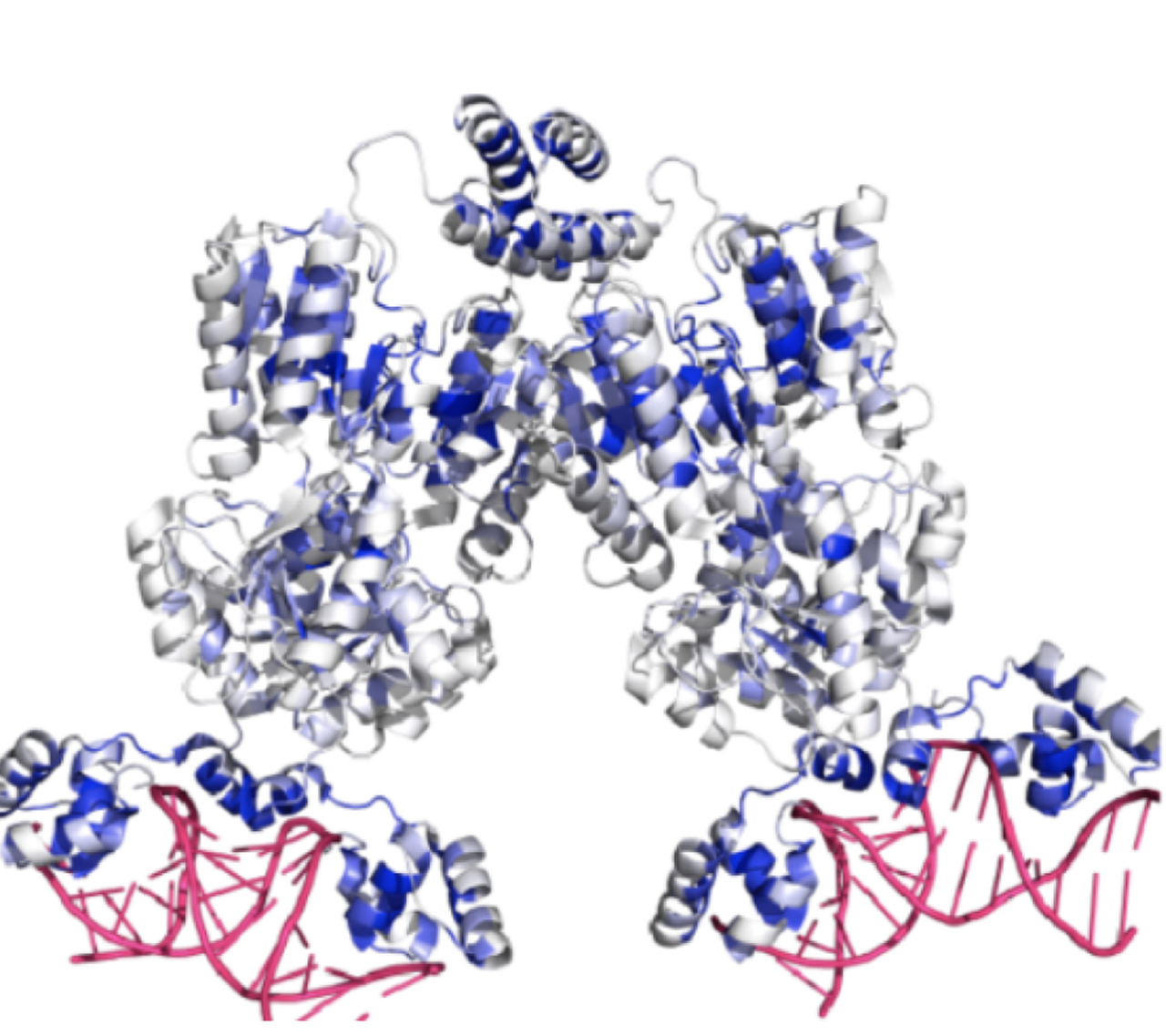
We have also explored deep mutational analyses of transcriptional repression mediated by the LacI protein. Repression can be conditionally toggled with a small molecule inducer that physically alters the shape of LacI to release DNA and allow productive transcription. Millions of single, double, and higher-order combinations of amino acid substitutions were generated and experimentally tested to dissect the functional consequence of variation to the protein and improve the resolution of the protein’s central mechanisms of transcriptional control. We developed computational approaches based on molecular simulation, co-evolution, and advanced representation learning from large language models to effectively predict the consequence on transcriptional repression after changing the LacI protein. Improved understanding of the functional core of LacI advances the general understanding of transcriptional control and accelerates engineering of custom biosensors for research, medical, and biomanufacturing advances.
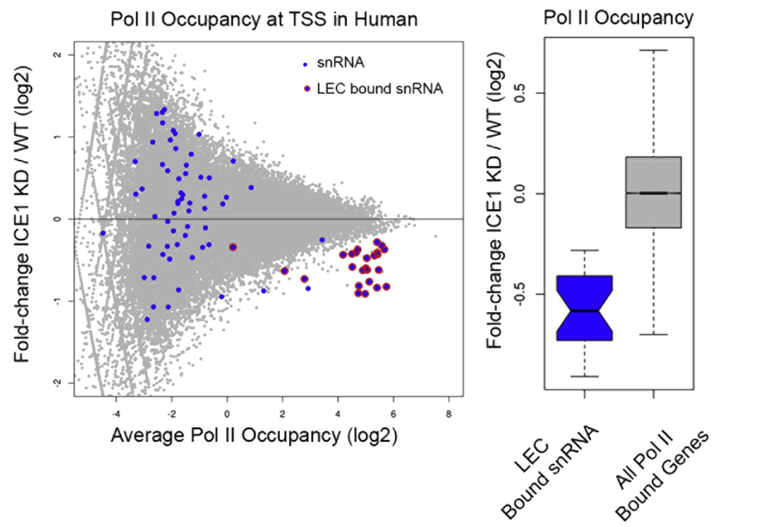
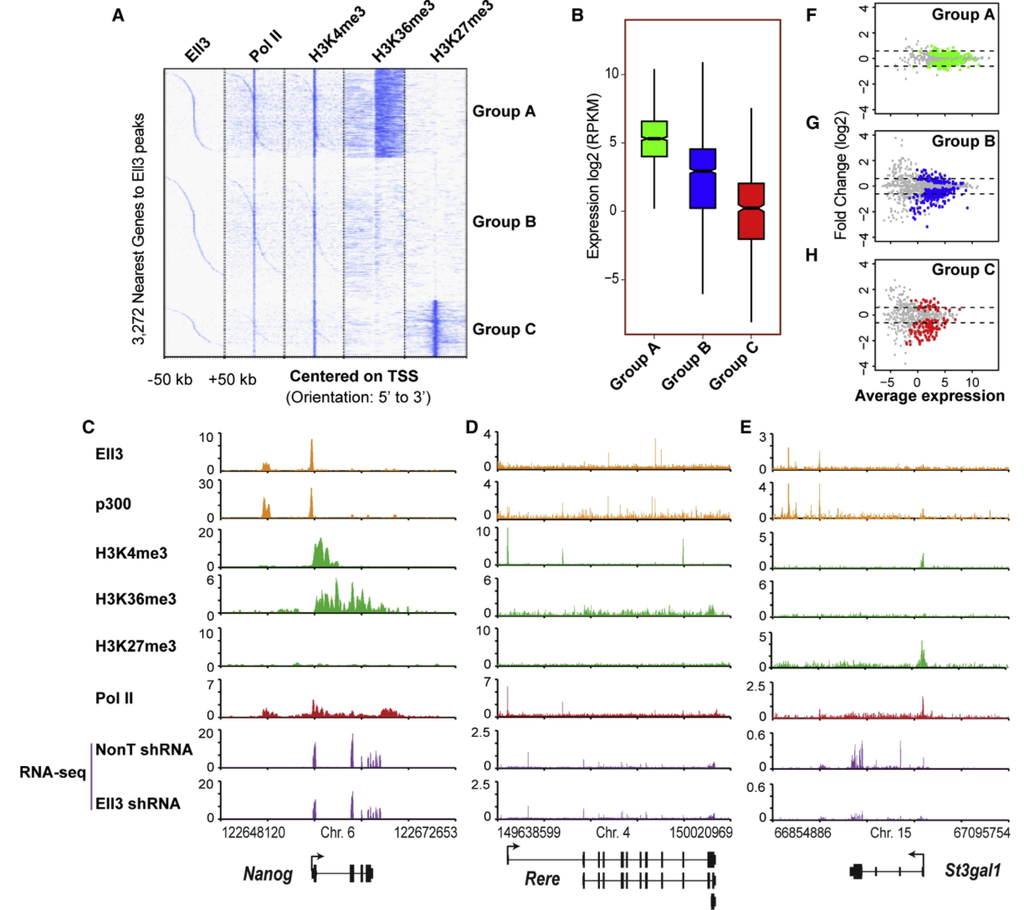
Additional research projects have included: binding site and target discovery of transcription factors controlling stem cell differentiation, chromatin modifications controlling the enhancer state of DNA, whole genome assembly, and more.