Press Release
23 August 2023
Stowers scientists find evidence of unintended impacts from anti-cancer drugs
Could potentially improve success rate of cancer drug development
Read Article
News
Centromeres—specific regions of DNA typically located near the center of chromosomes—achieve a common core structure to ensure proper alignment and segregation of chromosomes during cell division.
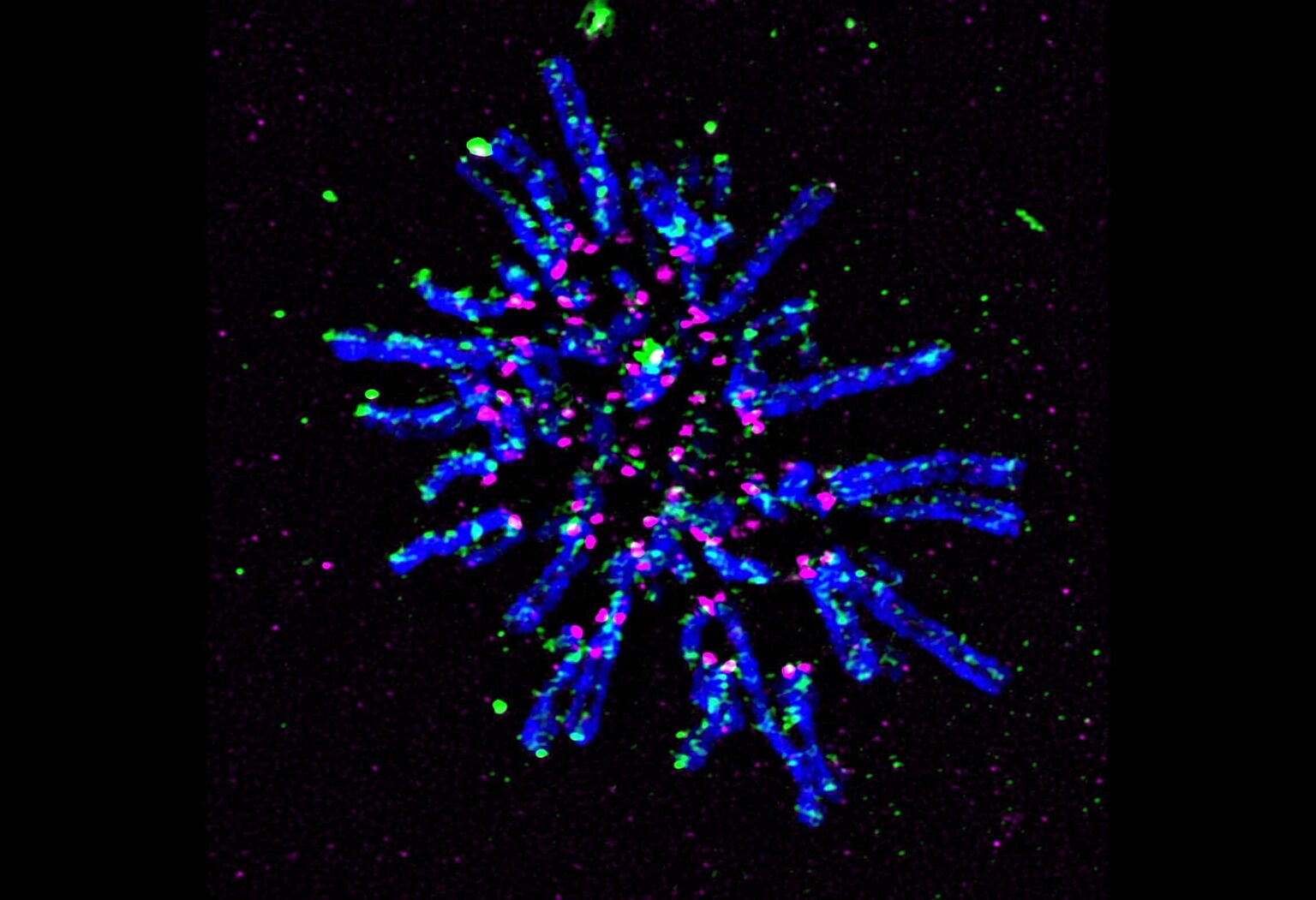
Super-resolution fluorescent microscopy image of chromosomes during cell division prior to aligning in preparation for separation. Chromosomes are in blue, a key protein that holds replicated chromosome together and facilitates DNA looping (cohesin) is in green, and the centromeric protein (CENP-A) is in magenta.
When cells divide, their chromosomes undergo astonishing feats where each one makes an identical copy and then drastically condenses into the characteristic ‘X’ that many people associate with chromosomes. How chromosomes are organized structurally at this stage is complicated yet critical for their ultimate separation and transfer into two cells from one.
New research published in Nature Communications on December 1, 2023 from the Stowers Institute for Medical Research explores the organization of human chromosomes at their centromeres—specific regions of DNA typically located near the center of chromosomes, corresponding to the midpoint of the ‘X.’ Centromeres play a crucial role in ensuring proper alignment and segregation of chromosomes during cell division. In humans, a key aspect of centromere structural organization has remained a mystery until now.
Understanding how centromeres are maintained has potential health implications. Disruptions in the relationship between centromere structure and how it functions may predispose certain people to chromosome instability and cancer.
The study, led by Postdoctoral Researcher Ayantika Sen Gupta, Ph.D., in the lab of Investigator Jennifer Gerton, Ph.D., found that despite widespread variability in the length and sequence of DNA in centromeres, they achieve a common structure to carry out their common function. The common structure allows for variable looping of DNA to keep it organized, similar to how neatly looping the long electrical cord of a hair dryer or a vacuum cleaner keeps it untangled.
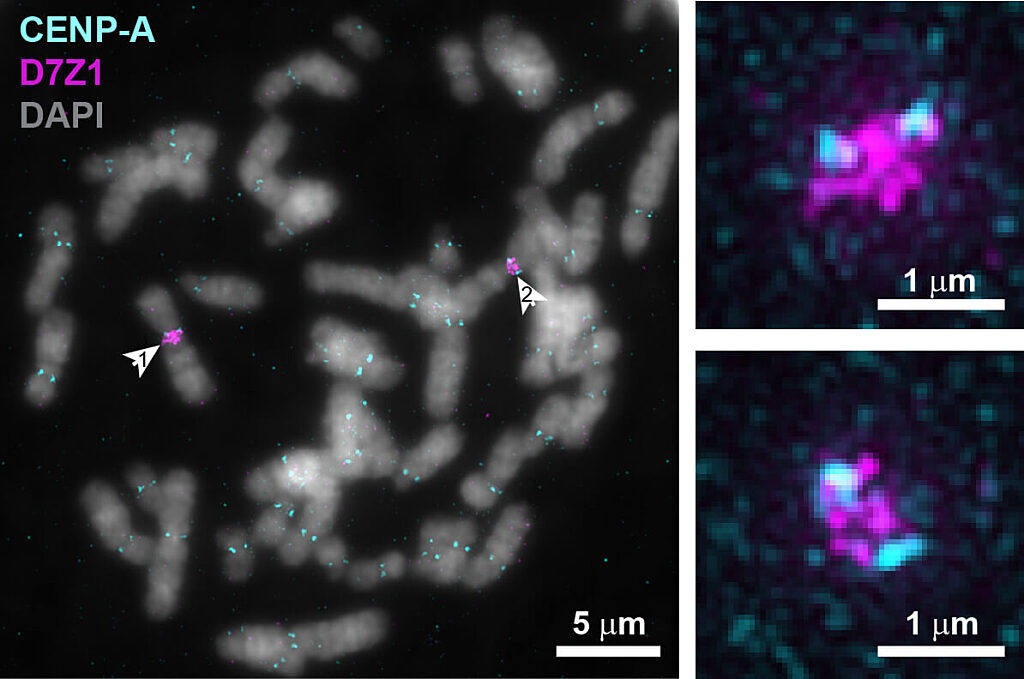
Fluorescent microscopy images of chromosomes during cell division with centromeric protein CENP-A in cyan and centromeric DNA in magenta. Arrows in left image point to a pair of chromosome 7s, with zoomed images on the right, indicating that an individual’s centromeric DNA is variable even between pairs of the same chromosome.
“Known in the field as the ‘centromere paradox,’ the length and sequence of human centromeric DNA is highly variable between different chromosomes as well as between the same chromosome in different people, even though the function is similar,” explained Sen Gupta.
Yet chromosome separation during cell division is a robust process that rarely fails. During routine cell division, or mitosis, individual chromosomes are replicated and must be held together prior to separating into the newly forming daughter cells. This chromosome cohesion is accomplished with a protein complex descriptively named “cohesin.”
Next, rod-like structures from each side of the dividing cell attach to the chromosomes and eventually pull them apart. However, the rods must attach at the right place and in the right orientation at the centromere, which can take several tries. These cycles of attachment and detachment appear as DNA stretching.
“The stretching observation has been known for many years,” said Sen Gupta. “You can think about stretching as a quality control mechanism. If a rod attaches to the wrong part of the centromere, it must detach and reattach correctly so that chromosomes can be distributed correctly to daughter cells.”
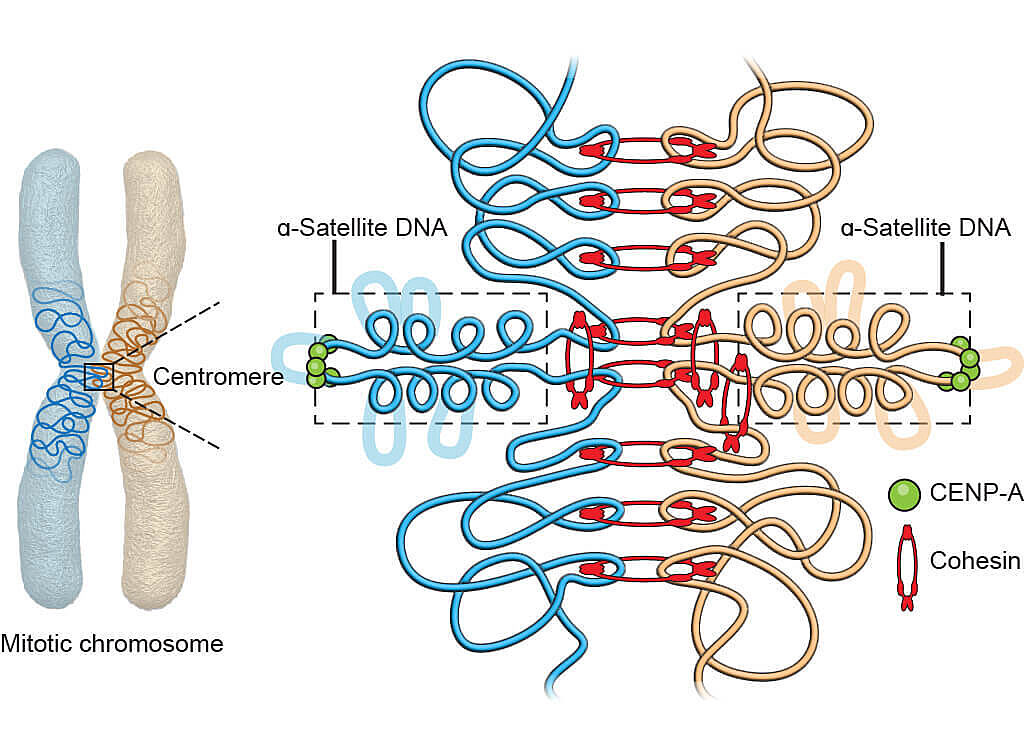
Graphical illustration of the 3D geometry model for the core centromere structure.
However, scientists have observed that the stretching motions far exceed the rigid nature of cohesin’s grip.
“We know what causes this motion but didn’t have a good model to explain it,” said Gerton.
Using genomic and microscopy techniques to study centromere components and structure in human cell lines, Sen Gupta developed a new model that helps explain the paradox. Cohesin attaches to the duplicated chromosomes tightly on either side of the region that undergoes stretching, allowing stretchy DNA to extend and then loop up when retracted, like the organized looping of long electrical cords.
Sen Gupta was able to show that several key protein complexes bestow a geometry that allows the formation of a common core centromere structure. Depending on centromere DNA variability, the core structure can accommodate a variable number of loops.
Next, the team plans to address whether there are implications connected to centromere variability. Even if centromeres achieve the same structure, their variability in sequence and length may imply different efficiencies of chromosome segregation during cell division. This question of how variability comes into play may indicate whether an individual is predisposed to chromosome instability, one hallmark of cancer.
“If you investigate these different centromere variations, are they all going to work equivalently or are some going to perform better than others?” asked Gerton. “We don’t know the answer to that yet but are hoping to find out.”
Additional authors include Chris Seidel, Ph.D., Dai Tsuchiya, Ph.D., Sean McKinney, Ph.D., Zulin Yu, Ph.D., Sarah Smith, Ph.D., and Jay Unruh, Ph.D.
This work was funded by the National Cancer Institute of the National Institutes of Health (NIH) (award: R01CA266339) and by institutional support from the Stowers Institute for Medical Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Press Release

23 August 2023
Could potentially improve success rate of cancer drug development
Read Article
News

27 September 2023
Q&A with Stowers Postdoc Ayantika Sen Gupta: "Explore as many science career possibilities as you can with an open mind."
Read Article
News

10 May 2023
Collaboration with Stowers scientists reveals mechanism underlying a common chromosomal abnormality
Read Article