News

31 March 2022
Completing the sequence
Stowers Institute researchers help assemble the complete human genome
Read Article
News
Collaborative study helps enlighten a paradox regarding chromosome transmission, potentially leading to a greater understanding of how variations in these regions underlie genome instability and disease
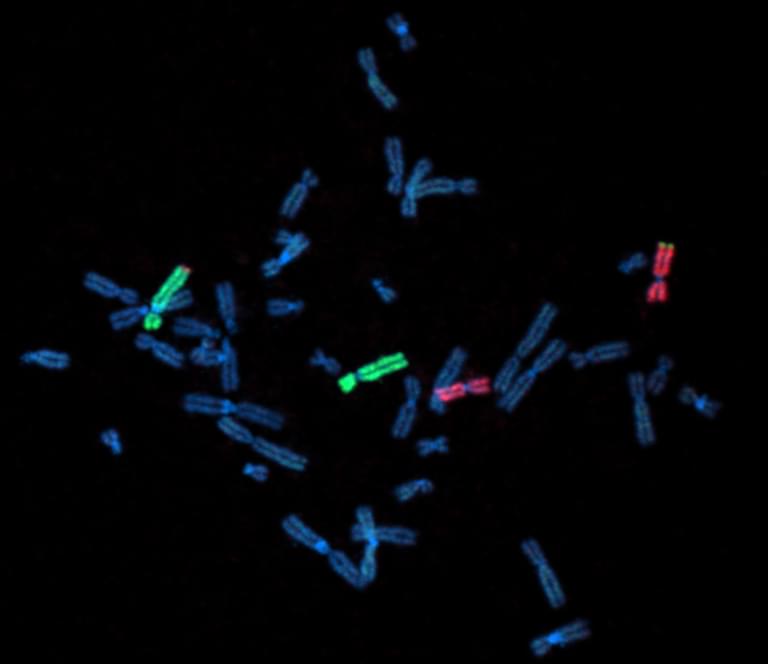
Fluorescent labeling of chromosomes in a newly sequenced human cell line. Chromosomes 4 and 11 are stained green and red, respectively.
When cells divide, each new cell needs a complete set of chromosomes. Centromeres, typically located near the center of chromosomes, provide attachment points to ensure that duplicated chromosomes are properly pulled to opposite sides of a dividing cell. However, the length and sequence of DNA at centromeres is highly variable among chromosomes, and even between the same chromosome in different people. This “centromere paradox” is a juxtaposition of centromere function with structure—the essential role of centromeres with their variable and rapidly evolving sequences.
A research study published online April 3, 2024, in Nature, led by Glennis Logsdon, Ph.D., and Evan Eichler, Ph.D., from the University of Washington with contributions from the lab of Stowers Institute Investigator Jennifer Gerton, Ph.D., presents the first comprehensive look at centromere variation in humans and non-human primates, shedding light on the centromere paradox. The study also lays groundwork for studying centromere variation, which could reveal insights underlying centromere mutation, genome instability, and human disease.
“Centromeres play a critical role in maintaining the integrity of the genome,” says Gerton. “If a cell loses the ability to transmit chromosomes accurately, it can have a major impact, sometimes leading to conditions like Down’s Syndrome or diseases like cancer.”
Centromeres are some of the most diverse and rapidly evolving regions in the genome. They are difficult to sequence because they contain highly repetitive stretches of DNA. In recent years, technological advances have greatly aided in their sequencing and assembly.
The authors compared two complete sets of human centromeres to establish a baseline for DNA sequence and structural variation. Then they compared this baseline to hundreds of other human centromere sequences. The team discovered substantial differences in the size and sequence organization of the centromeres and observed considerable variation in the location of attachment points within centromeres. The study provides estimates of the mutation rates of human centromeres underlying their variability and distinct evolutionary trajectories.
Next, the researchers sequenced a subset of chromosomes from chimpanzee, orangutan, and macaque genomes and compared them to human centromeres to better understand the rate of evolutionary changes over a 25-million-year period. These comparisons revealed substantial variation and mutation rates, suggesting that centromeres have largely independent evolutionary trajectories that shape their variation, with distinct structural features in centromeres characteristic to each species. This study helps provide an understanding for how these sequences evolve in primate lineages.
The Gerton Lab has a history of involvement with collaborative genome sequencing efforts. In 2022, Stowers scientists helped fill the gaps in the human genome that remained after the landmark DNA sequencing efforts of the Human Genome Project two decades prior. In 2023, the Gerton Lab contributed to the assembly of the first complete human Y chromosome.
Like prior collaborations, the current study includes contributions from Tamara Potapova, Ph.D., a scientist in the Gerton Lab with expertise in imaging chromosomes under the microscope. Using fluorescent staining techniques, Potapova analyzed the variation in the study’s new cell line’s karyotype, or the number and physical arrangement of chromosomes.
“Having complete centromere assemblies for humans and closely related animal species expands our understanding of chromosome biology and evolution,” said Potapova. “These assemblies are available for any researcher to use in their studies, which helps the scientific community in a broad way.”
“Other recent work from our lab has uncovered how human centromeres, despite differences in sequence length, can achieve a common core structure by variable looping of DNA. This may be one explanation for the centromere paradox,” said Gerton. “Continuing to explore the molecular mechanisms underlying centromere structure and activity may one day yield important insights into human health.”
This work was funded in part by the Stowers Institute for Medical Research and the National Institutes of Health including a grant from the National Cancer Institute to Jennifer Gerton (award R01 CA266339). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
News

31 March 2022
Stowers Institute researchers help assemble the complete human genome
Read Article
News
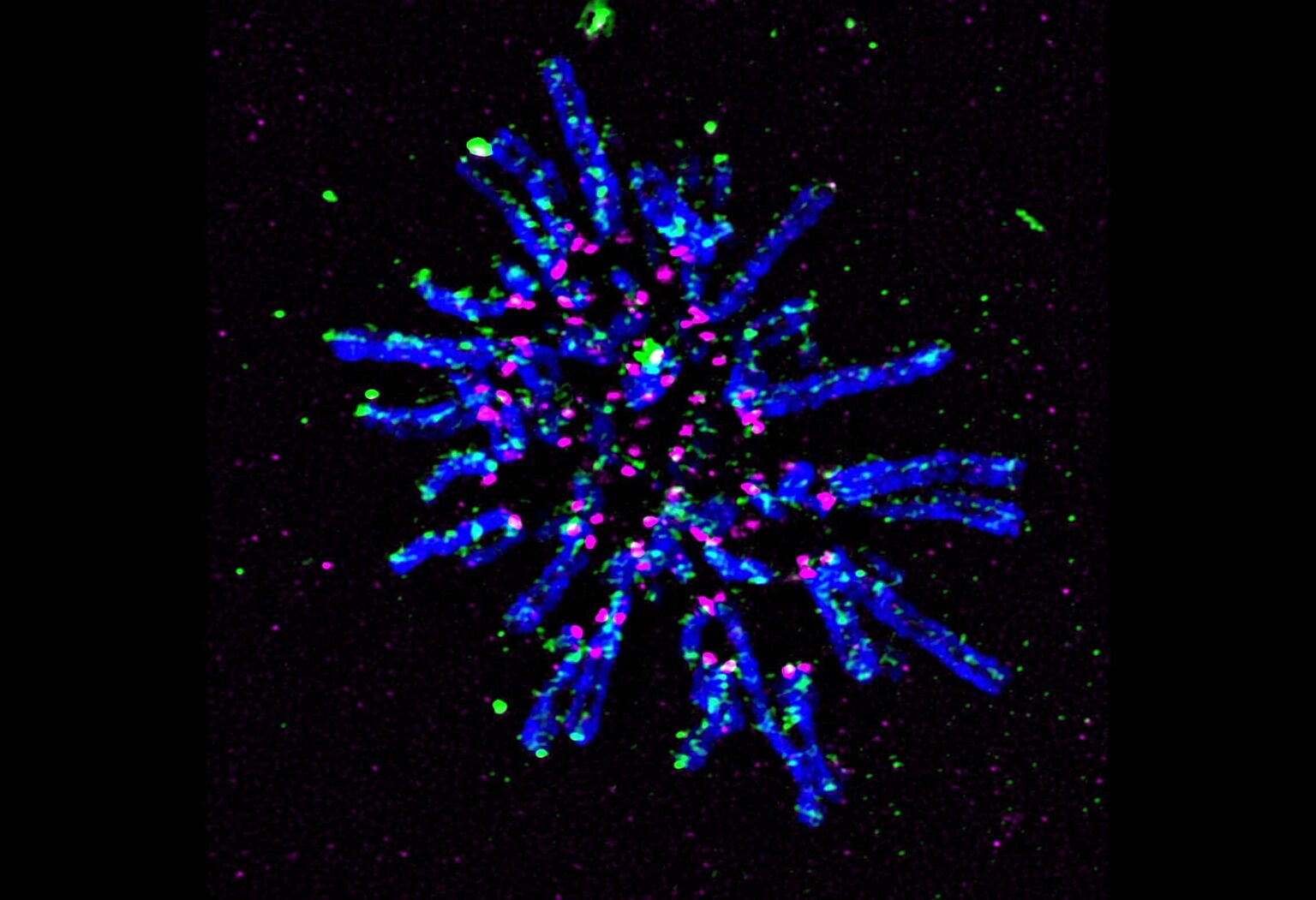
04 December 2023
Centromeres—specific regions of DNA typically located near the center of chromosomes—achieve a common core structure to ensure proper alignment and segregation of chromosomes during cell division.
Read Article
News

10 May 2023
Collaboration with Stowers scientists reveals mechanism underlying a common chromosomal abnormality
Read Article
News

24 April 2023
Foundational research at the Stowers Institute is leading to a greater understanding of many factors governing infertility with the goal of improving health, healthy outcomes, and providing hope for people facing this challenge.
Read Article