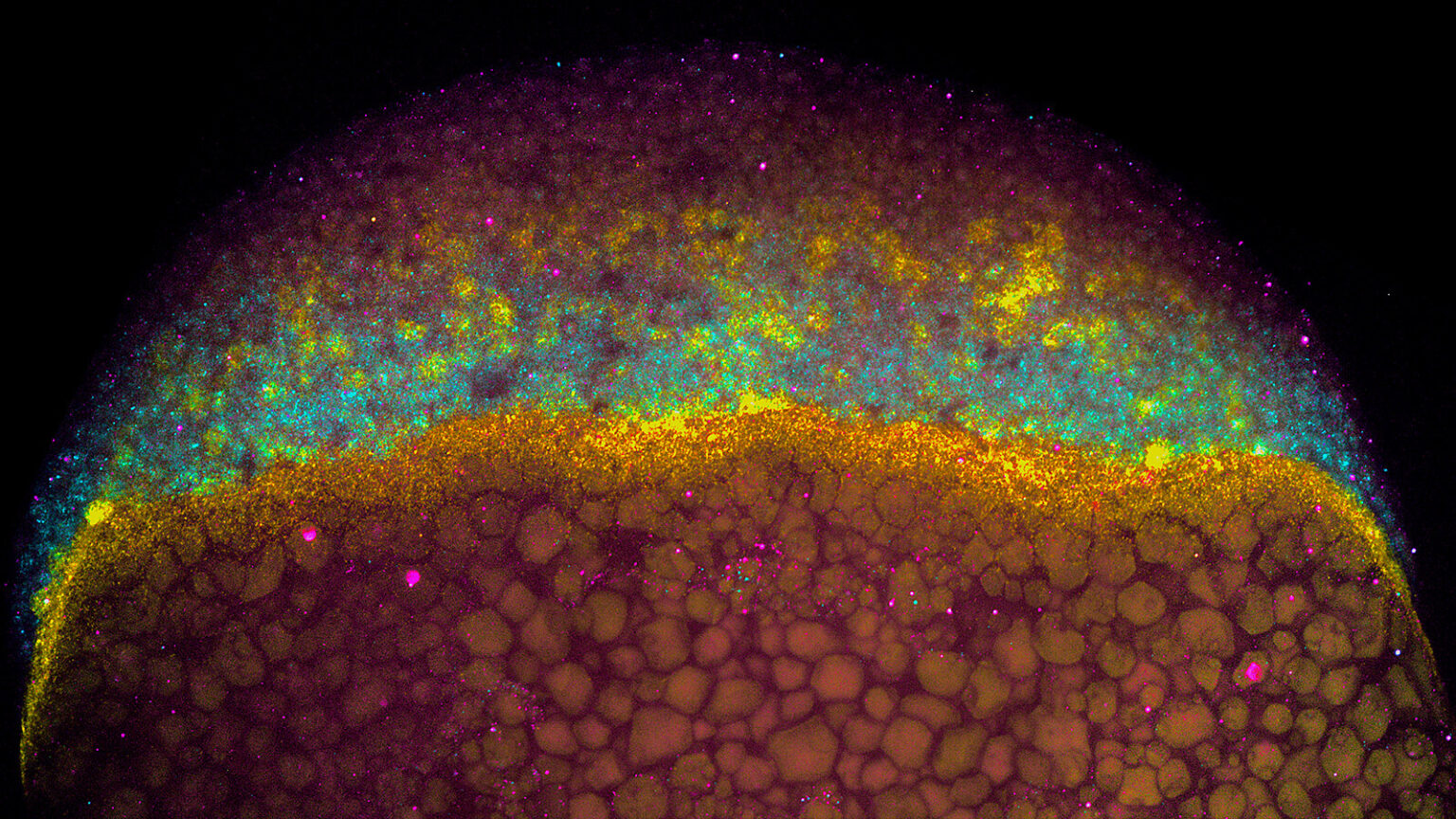
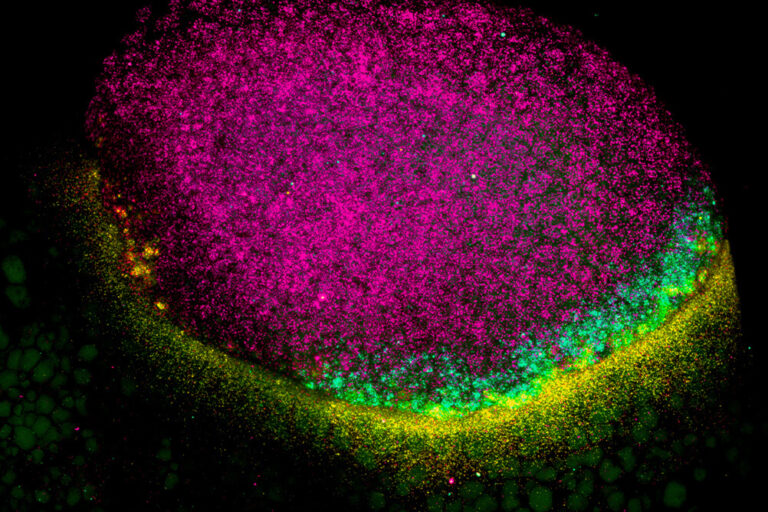
We study how mRNA stability and translation control gene expression during vertebrate development and viral infection. Although transcription was once considered the main determinant of gene output, we now know that how long an mRNA lasts and how efficiently it is translated play crucial roles in shaping protein levels. Our lab investigates the regulatory information encoded within mRNAs—such as sequence motifs, codons, and structural elements—that influence their stability and translation.
We focus on two major systems: zebrafish embryogenesis and viral infection. Zebrafish provide a powerful model to uncover how post-transcriptional regulation drives cell fate and early development, with lessons broadly conserved in humans. We also study how flaviviruses, such as dengue and Zika, rewire host gene expression, particularly at the level of translation, to ensure infection. Beyond these systems, we are exploring the hidden world of small open reading frames (sORFs) within noncoding regions, seeking to understand the novel microproteins they produce. Together, these efforts aim to reveal fundamental mechanisms of gene regulation with broad implications for development, physiology, and disease.