< All Technology Centers
Microscopy
The Microscopy center puts collaboration first: its full range of instrumentation, technical support staff, and image analysis tools are there to remove all barriers to the Stowers Institute’s research scientists when it comes to electron or light microscopy.
Overview of Services
Research Services
The Stowers Microscopy Center helps advance research at the institute by collaborating with lab members on scientific projects, maintaining a wide variety of microscopes both in the center and throughout the institute, and adapting or developing new technologies in anticipation of future imaging needs. The facility’s resources include electron microscopy, light microscopy, and computational resources. Our multidisciplinary team of physicists, software specialists, chemists, and biologists accelerate the pace of research with experimental design, execution, and analysis.
Technologies
- Zeiss Merlin SEM with Gatan 3View
- FEI Talos F200C TEM
- FEI Tecnai 120kV TEM
- Hitachi TM4000 SEM
- Zeiss Elyra 7 with Lattice SIM
- Leica SP8 STED with Falcon Lifetime
- Zeiss LSM 980 with AiryScan and OPO
- Nikon Ti Eclipse with CSU-W1 Spinning Disk and PLW20 Well Plate Loader
- Nikon Ti2 with CSU-W1 Spinning Disk and Opti Microscan FRAP Unit
- PerkinElmer Opera Phenix
- Olympus VS120 Slide Scanner
- Zeiss LSM 800
- Zeiss LSM 700
- Zeiss PALM Microbeam dissection microscope
- EM labeling techniques: Immuno-gold and APEX2
- 3D EM imaging: 3View, ATUMtome, tomography, traditional serial sectioning
- Super-resolution techniques: SIM, STED, PALM, STORM
- Protein interaction techniques: FRET, FRAP, FCS, FCCS
- Robotic imaging platforms and software for automated imaging
- Laser microdissection, ablation, and heat stimulation
- Selective Plane Illumination Microscopy (SPIM)
- Micro-fabrication and macro imaging
Software & Computing
- Fiji, IMOD, Imaris, Amira, Blender, napari, and various python libraries
- Deep learning using DeepFiji and Tensorflow
- 10 High performance workstations
- 2 Workstations with Deep Learning GPUs
Team Contact
Kexi Yi
Director of Microscopy
Stowers Institute for Medical Research
Kexi Yi joined the Stowers Institute for Medical Research in 2009 as a research specialist in the lab of former Investigator Rong Li, Ph.D. He joined the Microscopy Technology Center in 2015 and was promoted to Head of Electron Microscopy in 2023.

Team Contact
Zulin Yu
Head of Light Microscopy
Stowers Institute for Medical Research
Zulin Yu joined the Stowers Institute for Medical Research in 2008 as a postdoctoral researcher before joining the Microscopy Technology Center in 2010. Yu was appointed to manager of Light Microscopy in 2021. In 2023, he was promoted to Head of Light Microscopy.

Team Contact
Steph Nowotarski
Head of Electron Microscopy
Stowers Institute for Medical Research

Our Microscopy Suite
Seeing is believing – being able to find, track, or examine interactions is more important than ever. Our extensive array of microscopes allow Stowers researchers to tease apart details from samples of all sizes: from the molecular level up to whole animals.
Super-resolution
With the Zeiss Elyra 7 Lattice SIM, 60 nm optical resolution is achievable, extending sub-organelle or meiotic DNA structure imaging to up to 80 mm deep. For smaller samples the Leica SP8 STED can push the resolution limits even further to 40nm or better.

PerkinElmer Opera Phenix
High-speed imaging
The center automates the imaging of organisms from whole animals down to single cells for hundreds or thousands of samples with a robot-equipped Nikon spinning disk and a Perkin Elmer Opera Phenix.

Zeiss LSM 780
Multiplex and FRET Imaging
More colors equals more information. Using the Zeiss LSM 780 or the Leica SP8 with Falcon Lifetime we can separate up to 8 colors from one another, expanding researchers’ options for finding just the right target. With fluorescence lifetime imaging, looking at protein interactions with FRET is a snapshot away.
3-Dimensional Electron Microscopy
SEM imaging and reconstructing serial sections becomes routine with the Zeiss Merlin SEM. The 3View upgrade by Gatan does away with cutting artifacts by imaging the surface of the block face and cutting afterwards.

Talos 200C Transmission Electron Microscope
Ultimate Resolution
The new Talos 200C Transmission Electron Microscope gives resolution limited only by one’s ability to preserve a sample’s ultrastructure. The software and high beam energy allow users to acquire 3D tomography data in thicker sections for the first time. The accompanying cryo holder allows for screening of grids for single particle averaging analysis.
Software
Software has become as crucial to the enterprise of microscopy as confocal and electron microscopes. This is demanded by the advanced quantitation required in modern biology and by the sheer quantity of data acquired. In the Stowers Microscopy Center, we utilize open source platforms such as python, napari, and ImageJ, AI packages like Cellpose, Stardist, and Deeplabcut, and commercial software packages such as Imaris, Amira, and Dragonfly. Software developed for lab projects is made available at our github repos, and a suite of ImageJ plugins at research.stowers.org/imagejplugins. Please follow the instructions there to follow our Stowers Fiji update site.
Sean McKinney: https://github.com/jouyun
Chris Wood: https://github.com/cwood1967
Jay Unruh: https://github.com/jayunruh
News from the lab
News
21 December 2025
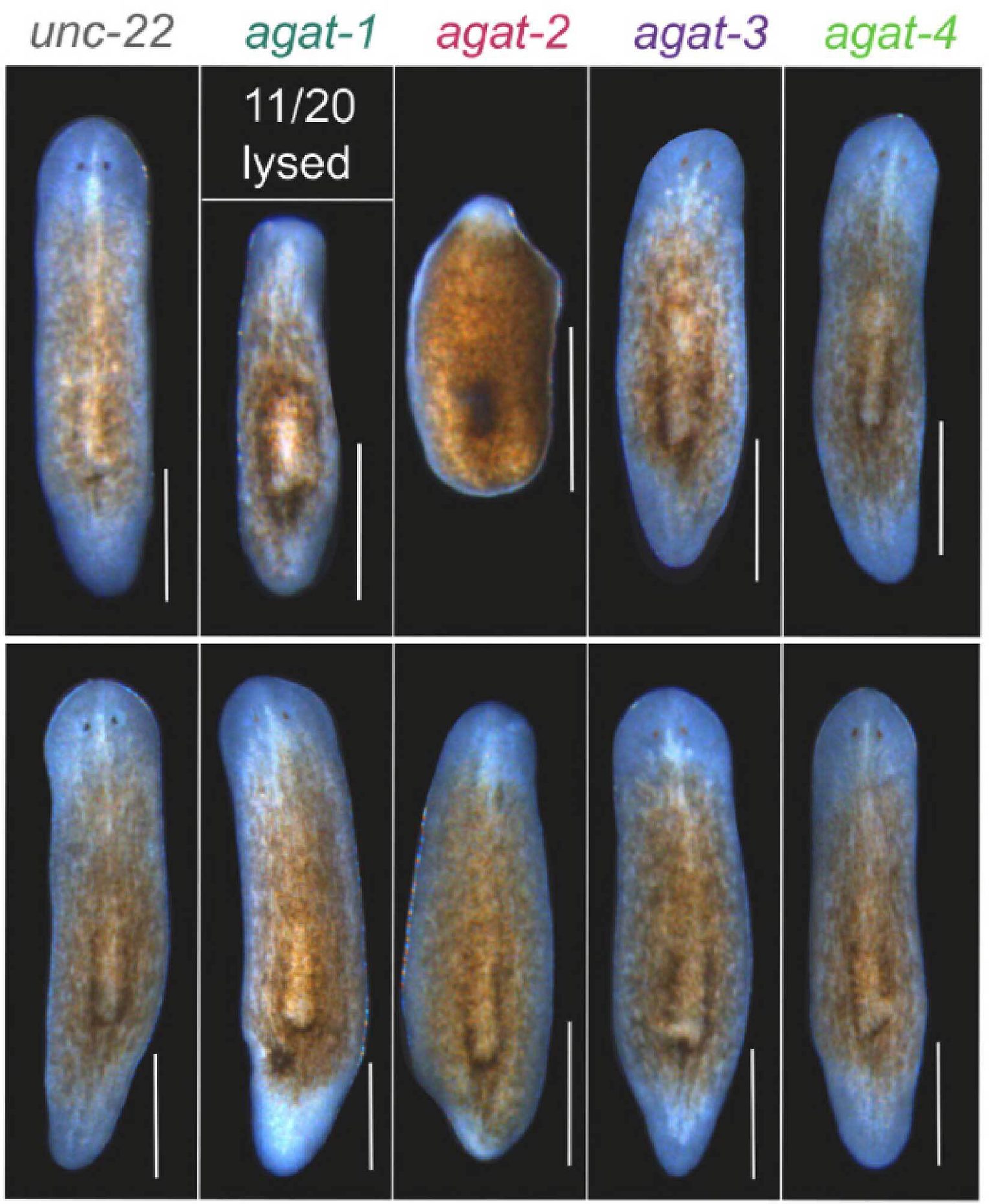
Support cells fuel flatworm regeneration
Stowers scientists discover how metabolically specialized support cells orchestrate whole-body regeneration, reshaping our understanding of tissue repair and regenerative medicine.
Read Article
News
15 April 2025
Light sheet microscopy: A decade-long journey from DIY innovation to cutting-edge imaging
A look at the technology that provides researchers with deeper insights into complex biological systems.
Read Article
News

07 March 2025
Road to Research: Q&A with Jeff Lange, Senior Research Scientist
"I hope to inspire early-career researchers to be curious and, in turn, have a far-reaching impact on humanity and human health as a whole."
Read Article
Featured Publications
wtf4 meiotic driver utilizes controlled protein aggregation to generate selective cell death
Nuckolls NL, Mok AC, Lange JJ, Yi K, Kandola TS, Hunn AM, McCroskey S, Snyder JL, Bravo Nunez MA, McClain M, McKinney SA, Wood C, Halfmann R, Zanders SE. eLife. 2020;9:e55694. doi: 55610.57554/eLife.55694.
Wnt and TGF-β coordinate growth and patterning to regulate size-dependent behavior
Arnold CP, Benham-Pyle BW, Lange JJ, Wood CJ, Sánchez Alvarado A. Nature. 2019;572:655-659.
Yeast centrosome components form a noncanonical LINC complex at the nuclear envelope insertion site.
Chen J, Gardner JM, Yu Z, Smith SE, McKinney S, Slaughter BD, Unruh JR, Jaspersen SL. J Cell Biol. 2019;218:1478-1490.
Hughes SE, Hemenway E, Guo F, Yi K, Yu Z, Hawley RS. PLoS Genet. 2019;15:e1008161. doi: 10.1371/journal.pgen.1008161.
Zeng A, Li H, Guo L, Gao X, McKinney S, Wang Y, Yu Z, Park J, Semerad C, Ross E, Cheng LC, Davies E, Lei K, Wang W, Perera A, Hall K, Peak A, Box A, Sánchez Alvarado A. Cell. 2018;173:1593-1608.e20.
An axial Hox code controls axial patterning and tissue segmentation in Nematostella vectensis
He S, Del Viso F, Chen CY, Ikmi A, Kroesen AE, Gibson MC. Science. 2018;361:1377-1380.