Zeitlinger Lab
Our goal is to understand the cis-regulatory code embedded in DNA that instructs gene regulation during development.
Welcome to the Zeitlinger Lab!
Our goal is to understand the cis-regulatory code embedded in DNA that instructs gene regulation during development. Using mouse and Drosophila embryos as model system, we study this fundamental problem with cutting-edge neural network models trained on high-resolution genomics sequencing assays, interpreted in the light of classical mechanistic studies.
What is the cis-regulatory code?
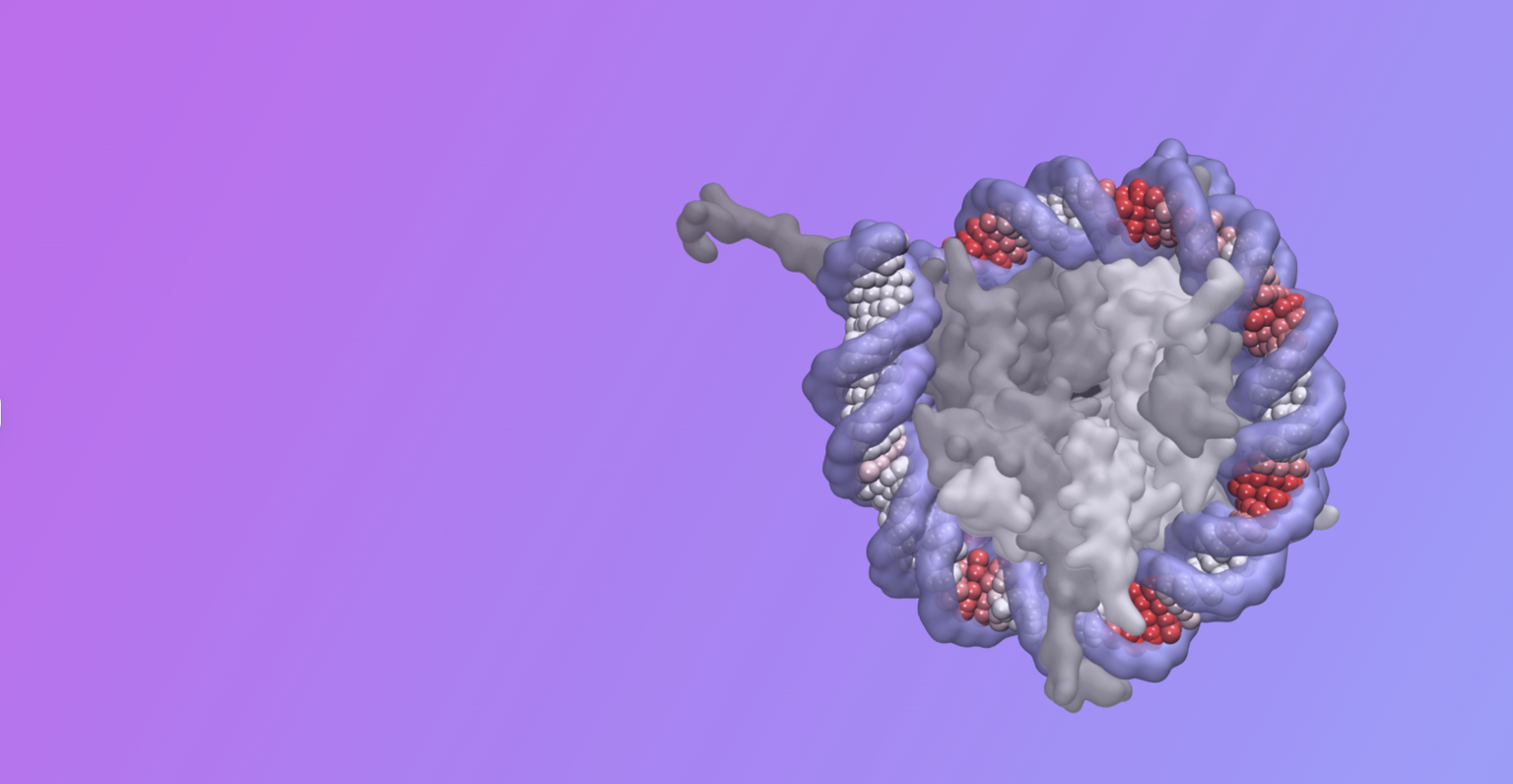
Developing embryos need to regulate genes at the right time and place to form the intricate structures of the body plan. These gene cis-regulatory instructions are encoded in DNA sequences called enhancers. They can be measured by a variety of assays, yet we cannot identify their function from sequence alone. Decades of research have focused on understanding the mechanisms by which transcription factors read out cis-regulatory sequences and regulate gene transcription, but this knowledge is insufficient to make accurate predictions about when and where cis-regulatory sequences instruct gene regulation in the embryo. If we could decipher the cis-regulatory code, this would unlock an enormous amount of information encoded in the human genome, much of which influences our disease disposition and response to treatments.
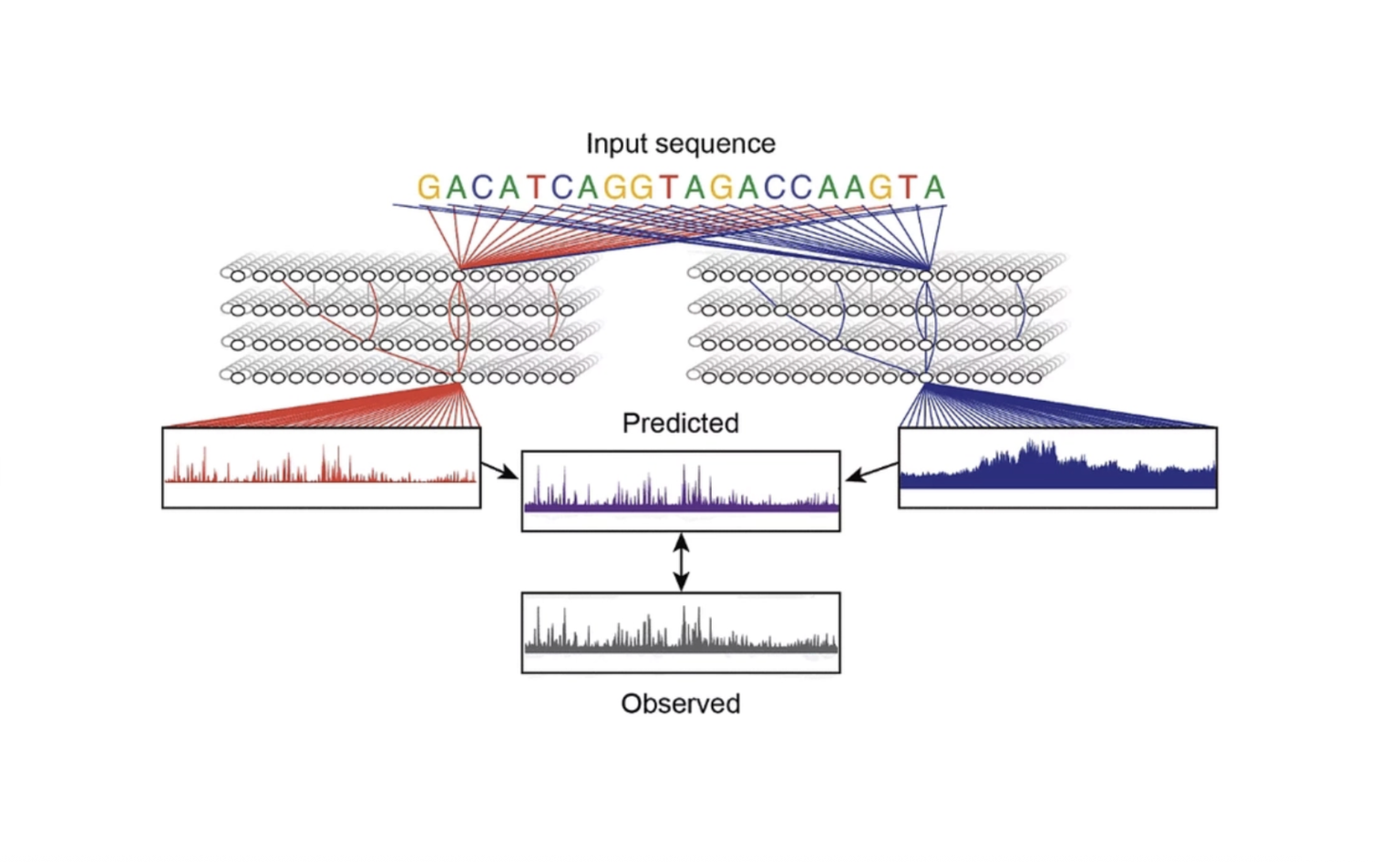
We have shown that neural networks trained on high-resolution transcription factor binding data make highly accurate predictions and learn cis-regulatory rules consistent with mechanistic studies. At the same time, they also reveal novel unexpected patterns that can be experimentally validated and inform models of gene regulation. We are expanding this approach to many cell types and genomic assays, including chromatin accessibility, nucleosome occupancy, histone modifications, 3D chromatin organization, RNA polymerase II initiation and pausing, and nascent transcripts. The goal is to learn the sequence rules that drive each gene regulatory process such that we can eventually build models that read the entire cis-regulatory code for many cell types.
What questions are we asking?
Learn more about our research
