
Our research
How do transcription factors bind cooperatively to DNA? Our models suggest that transcription factors have two modes of cooperativity, one through strictly spaced motifs and one through motifs with soft syntax within nucleosome distance. To further understand these mechanisms, we combine the results from our neural network model with MD simulations and in vivo CRISPR experiments.
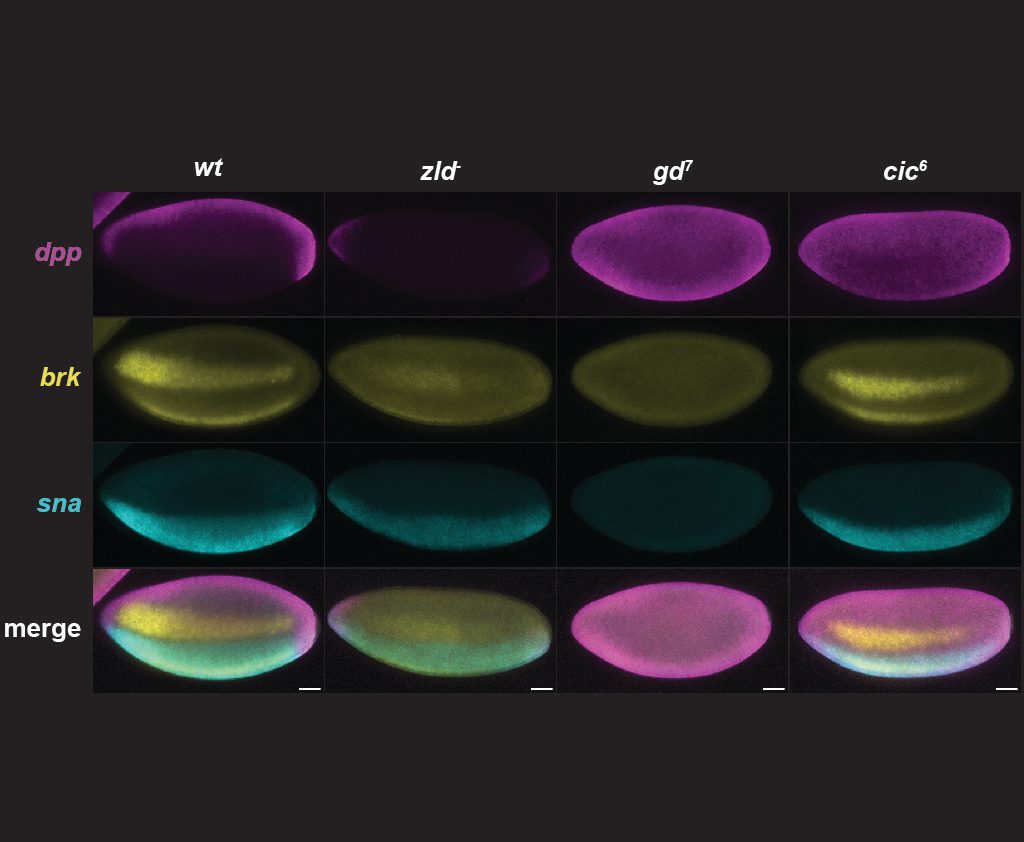
How do transcription factors overcome the nucleosome barrier and make DNA accessible in chromatin? Our models suggest transcription factors pioneer chromatin proportional to motif affinity and in many cases act on nucleosomes cooperatively. We are currently investigating thermodynamic models for this type of cooperativity, how it affects transcription factor binding and how it might change the properties of enhancers.
Questions we're asking
Explore our areas of research below

How do nucleosomes contribute to the cis-regulatory code?
We have created novel neural network models that predict the genome-wide occupancy of nucleosomes without the experimental bias imparted by MNase. Leveraging these models, we study the motifs that position or deplete nucleosomes and how they affect gene regulation. We also investigate how sequences not corresponding to transcription factor motifs might influence gene regulatory processes.
How do transcription factors mediate gene activation?
We take a variety of approaches to fill the remaining gaps in how the right combination of transcription factors form active enhancers, which induce nearby promoters to transcribe a gene. This includes interpreting models of histone modifications and enhancer RNA found at active enhancers, and analyzing high-resolution data of RNA polymerase II initiation and pausing
How do we adopt and develop genomics technology that best informs the cis-regulatory code across many cell types?
We have previously developed ChIP-nexus technology to map transcription factors at the highest possible resolution in vivo. We are currently expanding our capabilities for single-cell and single-molecule approaches.
Explore the Lab
Check out the videos below to learn more about our work.
Let’s take a #LookInTheLab! What does the Zeitlinger Lab study? Kaelan Brennan, a Stowers Graduate School predoc in the lab, explains their work on gene regulation using multiple systems.
The cis-regulatory code that instructs gene regulation, also known as the genome’s second code, is a fundamentally unresolved problem. It is estimated that over 80% of disease-causing mutations in the human genome are found in cis-regulatory regions, but since we cannot read the code, we cannot predict which genetic variants disrupt gene regulation. Recent progress in using neural networks for learning DNA sequence has however provided proof-of-principle that this complex cis-regulatory code can be learned. This new approach is fundamentally different from traditional methods in that it is an inverted learning paradigm.
With artificial intelligence (AI) poised to greatly accelerate the pace for novel discoveries in foundational biological research, the Stowers Institute launched the Office of Scientific Leadership AI Initiative, a new program designed to advance capabilities in machine learning and AI for addressing critical biological questions. Investigator Julia Zeitlinger, Ph.D., has been appointed to lead this effort and leverage cutting-edge computational techniques to accelerate scientific discoveries and drive innovation in biological research.
