News
18 November 2025
Decoding early development
Stowers Associate Investigator Ariel Bazzini, Ph.D., discusses a collaboration that uncovered a new mechanism guiding the earliest steps of life.
Read Article
M.S., Biology, National Institute of Agriculture Technology, Biotechnology Institute and University of Buenos Aires
Ph.D., Biology, National Institute of Agriculture Technology, Biotechnology Institute and University of Buenos Aires
Associate Investigator
Stowers Institute for Medical Research
Associate Professor
The Graduate School of the Stowers Institute for Medical Research
Associate Professor
Department of Cell Biology and Physiology, University of Kansas School of Medicine

Associate Investigator
Stowers Institute for Medical Research
Associate Professor
The Graduate School of the Stowers Institute for Medical Research
Associate Professor
Department of Cell Biology and Physiology, University of Kansas School of Medicine
We don’t just do science that we can do or that we feel comfortable doing. It’s about doing the science that we want to do.
Research Areas
Development and Regeneration, Genetics and Genomics, Molecular and Cell Biology
Courses Taught
Gene Expression: Transcription to translation; Laboratory Rotation; Thesis laboratory
Honors
2019-2021
Pew Innovation Fund
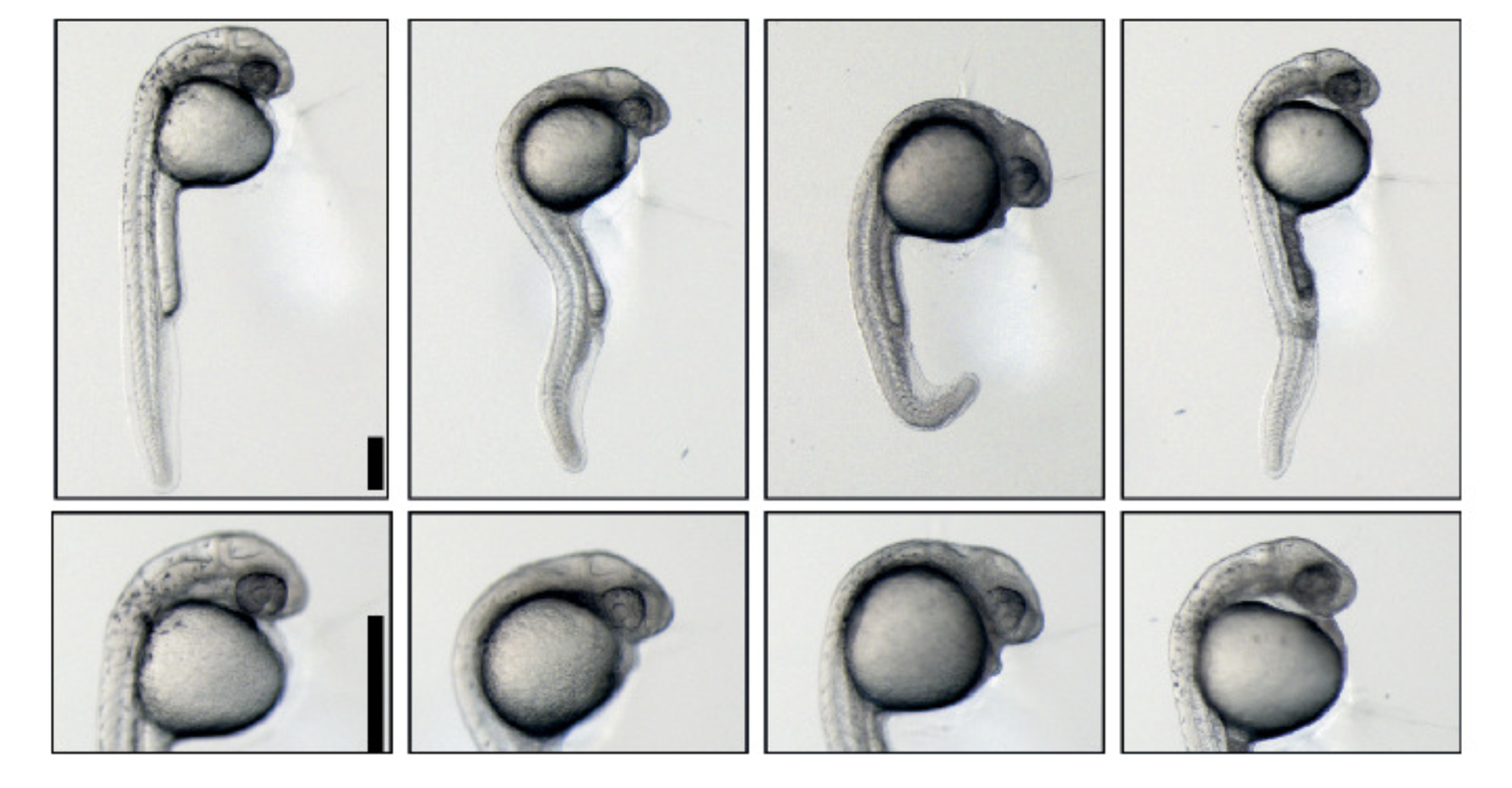
Ariel Bazzini, Ph.D., joined the Stowers Institute in 2016. His research focuses on gene regulation in development and disease and in unraveling the intricacies in RNA stability and translation.
Bazzini earned a Ph.D. in biology from the National Institute of Agriculture Technology, Biotechnology Institute and University of Buenos Aires. As a doctoral student studying molecular biology, he was fascinated by RNA, and especially messenger RNA (mRNA), a molecule well-known for translating the DNA code into the amino acids that make up proteins.
Bazzini first studied RNA in depth while investigating how viruses affect gene expression in plants during his doctoral studies at the Institute of Biotechnology in Argentina’s National Institute of Agricultural Technology and the University of Buenos Aires. To expand expertise in animal genetics, he took a postdoctoral fellowship position at Yale University in the lab of Antonio Giraldez, Ph.D., who was researching microRNA in zebrafish. Bazzini successfully adapted the ribosome profiling technique (for the first time in a whole embryo) to measure translation in vivo to understand the large range of translation levels between mRNAs in a cell. He initially used this state-of-the-art technique to demonstrate that miRNAs sequentially prevent translation and subsequently RNA decay during embryogenesis.
Using ribosome profiling to define the coding region of the entire zebrafish transcriptome enabled identification of small, translated coding regions in 5’ and 3’ “untranslated regions” (UTRs), as well as in genes that were annotated as non-coding. Later, he showed that coding regions in the 5’UTR are translational repressors.
Bazzini is also interested in understanding what defines the stability of the mRNAs. He discovered that codons, the three-nucleotide ‘words’ read by ribosomes, have a strong effect on the stability of maternal mRNA and thus contain cis-regulatory information that extends far beyond the amino acids they encode. This represents a paradigm shift in how mRNA stability is viewed.
News

18 November 2025
Stowers Associate Investigator Ariel Bazzini, Ph.D., discusses a collaboration that uncovered a new mechanism guiding the earliest steps of life.
Read Article
News
12 August 2025
Horacio Martín Pallarés, Ph.D., in the lab of Ariel Bazzini, Ph.D., a 2010 Pew Fellow, plans to bring his research on viral epidemics to aid his home country of Argentina
Read Article
News

09 May 2025
"For me, joining the Stowers Institute was a chance for me and my team to follow any research idea we’re curious about."
Read Article