Research
Research
Research Interests
The eukaryotic ribosome is a complex molecular machine responsible for the translation of mRNA to protein. Over the years we have gained detailed knowledge of ribosome structure, function, and biogenesis; however, a major unanswered question in the translation field is how cells monitor the integrity of the ribosome itself. Alterations in ribosome structure and function have been associated with diseases such as neurodegeneration, cancer and ribosomopathies. Indeed, mutations, environmental stress, or mistakes during assembly can lead to malfunctioning ribosomes that need to be detected and marked for degradation to maintain translation fidelity. My group capitalizes on the recent advances in quantitative mass spectrometry, functional genomics methods, and CRISPR-mediated genome editing to study the molecular mechanisms of ribosome surveillance.
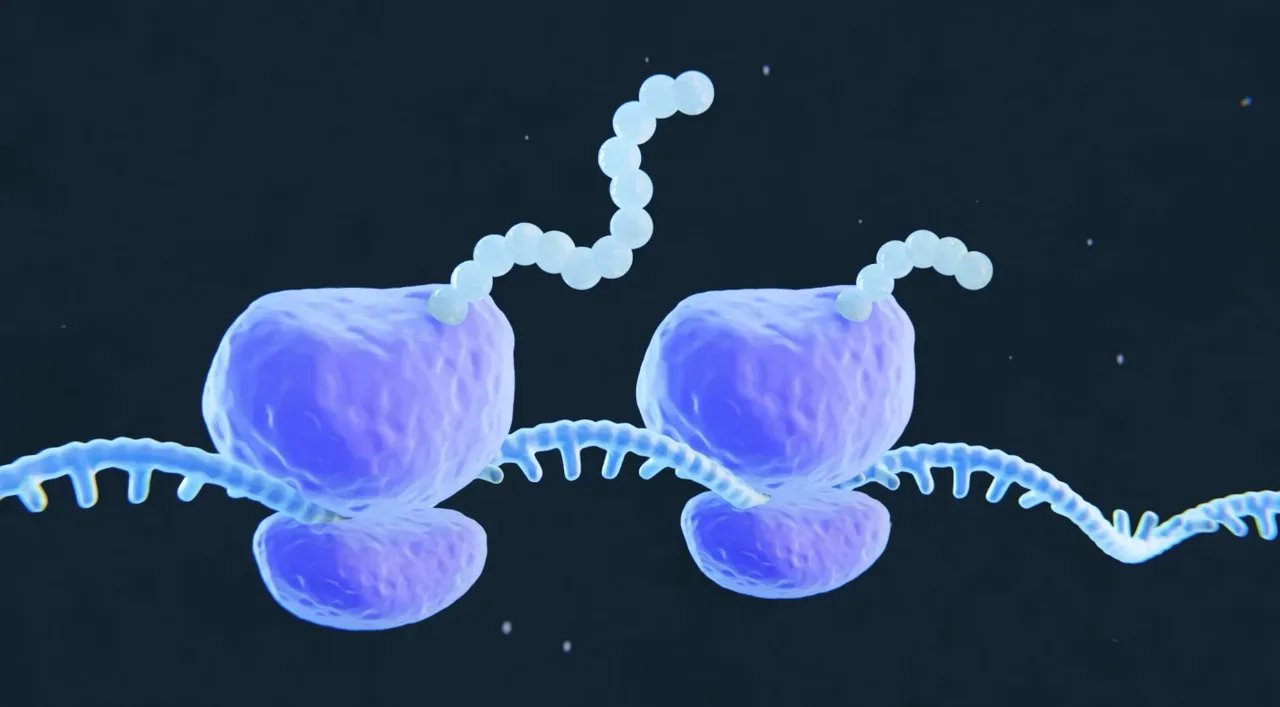
Ribosome quality control
The ribosome is one of the most complex and long-lived machines in the cell and its many components are subject to both mechanical and chemical assaults. Accumulated damage can render the ribosome elongation incompetent, causing it to stall on messages. We are interested in identifying the molecular pathways that cope with such damaged ribosomes.
Image credit: Caitlin Rausch, Warbler Creative

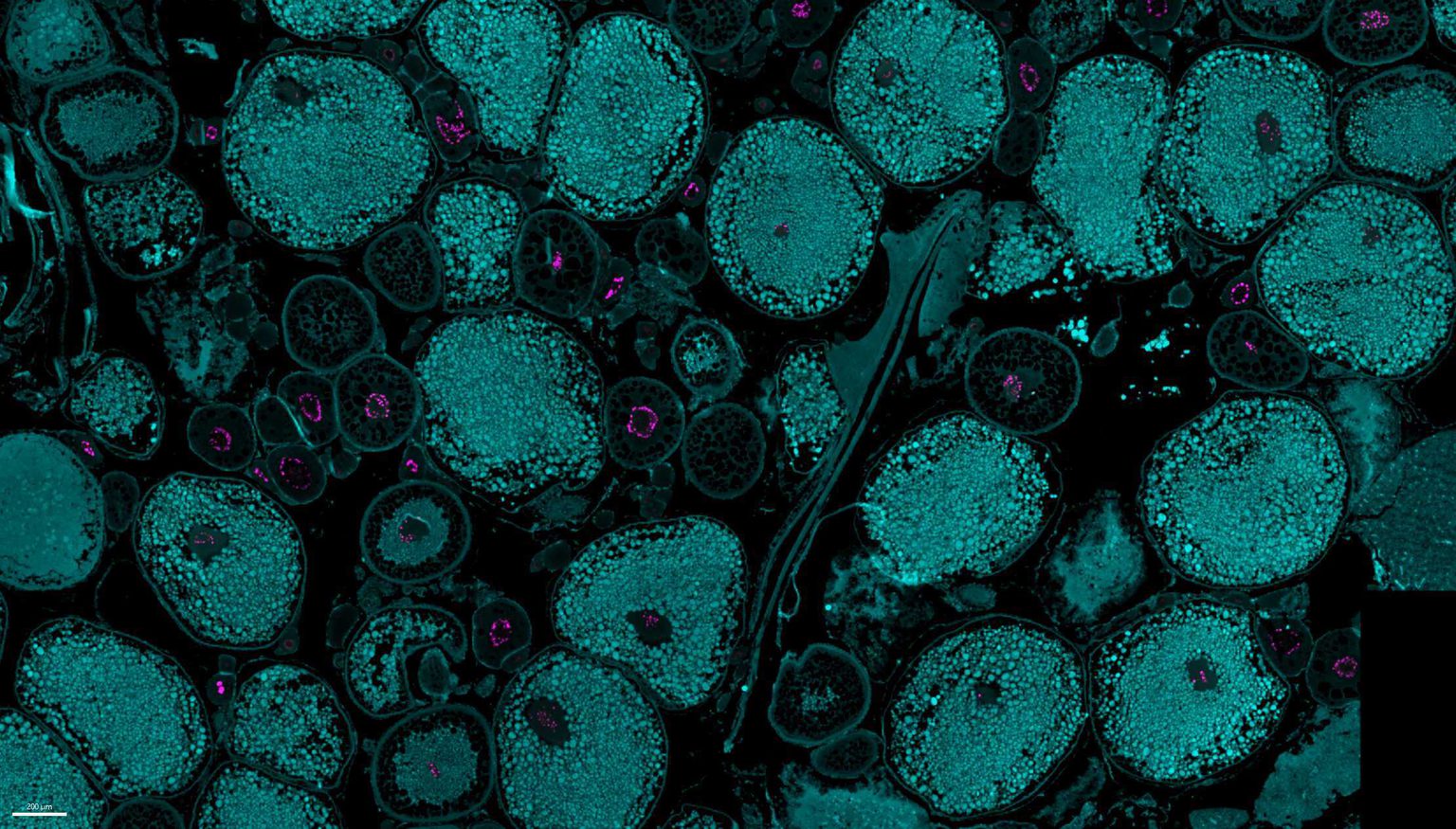
Ribosome assembly and oogenesis
Oocytes are among the largest cells in nature, tasked with supporting early development before the onset of zygotic transcription. To meet this challenge, eggs accumulate an extraordinary number of ribosomes—often billions—before fertilization. This rapid and massive ribosome production requires unique adaptations in ribosome biogenesis pathways, including amplified rDNA transcription and highly specialized RNA processing and assembly mechanisms. We use zebrafish as a model to investigate how oocytes build their ribosome pool, asking how these specialized pathways are regulated and whether they produce ribosomes with unique properties tailored for early embryogenesis.
Ribosome heterogeneity
The eukaryotic ribosome is composed of four ribosomal RNAs (rRNAs) and approximately 80 ribosomal proteins. For decades, it was assumed that organisms build structurally identical ribosomes across tissues, developmental stages, and environmental conditions, including during disease initiation and progression. However, this view has been overturned by discoveries showing changes in both rRNA and protein composition. A striking example comes from zebrafish , where two distinct ribosome populations—maternal and somatic—carry out protein synthesis during early embryogenesis. We are interested in understanding how such ribosome diversity shapes translation and drives early embryonic events.